Peroxisome Proliferator-Activated Receptor ÃÆà ½Ãâò/ÃÆà ½Ãâô Stimulation Induces the Differentiation of Human Alveolar Epithelial Progenitor Cell
Chihiro Ozawa, Michiko Horiguchi, Tomomi Akita, Mai Hirokawa, Yusuke Nakao, Kaori Abe and Chikamasa Yamashita
DOI10.21767/2577-0578.10011
Michiko Horiguchi*, Chikamasa Yamashita*, Chihiro Ozawa, Tomomi Akita, Mai Hirokawa,Yusuke Nakao, Kaori Abe
Department of Pharmaceutics and Drug Delivery, Faculty of Pharmaceutical Sciences, Tokyo University of Science, 2641 Yamazaki, Noda, Chiba 278-8510, Japan
- *Corresponding Authors:
- Michiko Horiguchi
Department of Pharmaceutics and Drug Delivery
Faculty of Pharmaceutical Sciences, Tokyo University of Science
2641 Yamazaki, Noda, Chiba 278-8510, Japan.
Tel: +81 4 7121 3691
Fax: +81 4 7121 3622
E-mail: horiguchim@rs.tus.ac.jp
- Chikamasa Yamashita
Department of Pharmaceutics and Drug Delivery
Faculty of Pharmaceutical Sciences, Tokyo University of Science
2641 Yamazaki, Noda, Chiba 278-8510, Japan.
Tel: +81 4 7121 3691
Fax: +81 4 7121 3622
E-mail: chikamasa_yamashita@rs.tus.ac.jp
Received date: August 26, 2015; Accepted date: February 10, 2016; Published date: February 19, 2016
Citation: Ozawa C (2015) Peroxisome Proliferator-Activated Receptor β/δ Stimulation Induces the Differentiation of Human Alveolar Epithelial Progenitor Cell. Insights Chest Dis. 1:11.
Abstract
Pulmonary emphysema is a disease in which alveoli in the lung are irreversibly destroyed, compromising lung function. All-trans-retinoic acid (ATRA) reportedly repairs the alveoli of emphysema model mice. In addition, ATRA can activate two nuclear receptors, nuclear retinoic acid receptor (RAR) and peroxisome proliferator-activated receptor (PPAR) β/δ. Both of these receptors induce differentiation in many different cell types. In this study, PPARβ/δ stimulation is shown to induce differentiation of human alveolar epithelial progenitor cells into alveolar type II epithelial (AT-II) cells. PPARβ/δ activation also stimulates the expression of adipose differentiation-related protein mRNA in human alveolar epithelial progenitor cells. These data suggest that PPARβ/δ selective agonists could differentiate human alveolar epithelial progenitor cells without cytotoxic of ATRA derived from stimulation to RAR.
Keywords
All-trans-retinoic acid; Cell differentiation; Lung; Peroxisome proliferator-activated receptor; adipose differentiation-related protein
Abbreviations
ADRP: Adipose Differentiation-Related Protein; AEPC: Alveolar Epithelial Progenitor Cell; ATRA: All-Trans-Retinoic Acid; GAPDH: Glyceraldehyde- 3-Phosphate Dehydrogenase; PPAR: Peroxisome Proliferator-Activated Receptor; RAR: Retinoic Acid Receptor; SP-A: Surfactant Protein A
Introduction
Chronic obstructive pulmonary disease (COPD), the fourth leading cause of death according to a report released by World Health Organization in 2005, is an increasing global health problem. COPD includes three pathological manifestations: chronic obstructive bronchitis, mucus plugging, and pulmonary emphysema. Pulmonary emphysema is a disease in which the lung’s gas-exchange structures, the alveoli, are irreversibly destroyed, causing the patient to die of compromised lung function unless a transplant is provided. The causes of COPD have not been clearly defined. However, many lipid mediators, inflammatory peptides, reactive oxygen and nitrogen species, chemokines, cytokines, and growth factors have been implicated in alveolar destruction [1].
Human alveolar epithelial progenitor cells (AEPCs) have been shown to self-renew. They also can differentiate into alveolar type II epithelial (AT-II) cells upon stimulation with a growth factor. Human AEPCs are purified from adult human lung using two selection procedures. The first is a negative selection that removes hematopoietic cells using an autoMACS Separator with anti-human CD45 antibody-coated microbeads. The second is a positive selection that picks out lung stem cells using FACSAria with anti-human pro-SP-C (an alveolar cell marker) and antihuman CD90 (a mesenchymal stem cell marker) antibodies [2].
Recently, we showed that all-trans-retinoic acid (ATRA), a metabolite of retinol (vitamin A), has the ability to induce the differentiation of human AEPCs to AT-II cells. AT-II cells have the ability to proliferate and differentiate into alveolar type I epithelial cells through the alveolar repair process. ATRA treatment also significantly improves the CT values of normal mice and elastasetreated mice, a mouse model of emphysema [3]. However, the detailed mechanisms underlying ATRA-induced differentiation of human AEPCs remain unclear.
ATRA can activate two nuclear receptors: nuclear retinoic acid receptor (RAR) and peroxisome proliferator-activated receptor (PPAR) β/δ [4]. RAR activation has several effects in cells, such as retinoic acid metabolism, cell cycle arrest, and mammary adenocarcinoma cell apoptosis [5] as well as acute promyelocytic leukemia cell differentiation [6,7]. Furthermore, PPARβ/δ activation causes many effects in cells, such as glucose and fatty acid metabolism [8], keratinocyte proliferation and differentiation [9,10], and adipocyte and sebocyte differentiation [11].
In this study, we assessed the contributions of nuclear receptors RAR and PPARβ/δ to the differentiation of human AEPCs using the selective RAR ligand TTNPB and the selective PPARβ/δ ligand GW0742. TTNPB can activate all subtypes of RARs, RARα, RARβ, and RARγ, and TTNPB has 1000-fold more potent as a teratogen than ATRA. It is also known that TTNPB is more stable than ATRA in minimum essential media containing FBS, and it seems to contribute to the potency of TTNPB [12,13]. And it is reported that GW0742 affects to PPARβ/δ 1000-fold much than those to PPARα and PPARγ [14,15].
Materials and Method
Reagents
4-[(E) -2- (5, 6, 7, 8-tetrahydro-5, 5, 8, 8-tetramethyl-2- naphthalenyl) -1-propenyl] benzoic acid (TTNPB) and [4- [[[2- [3-fluoro-4- (trifluoromethyl) phenyl] -4-methyl-5-thiazolyl] methyl] thio] -2-methyl phenoxy] -acetic acid (GW0742) were purchased from Enzo Life Sciences, Inc. (USA) and Cayman Chemical (USA) respectively. These chemicals were dissolved in dimethyl sulfoxide (DMSO) for biochemistry purchased from Wako pure chemicals (Japan) and stored in the dark at -20 °C before use. Antibodies against SP-A (cat. Sc-7699), actin (cat. sc-1616) and goat -HRP conjugated (cat. sc-2020) were obtained from Santa cruz Biotechnology Inc. (USA). Donkey anti-goat IgG-FITC (cat. AP180F) was purchased from EMD Millipore Corporation (USA). DAPI (4, 6-diamino-2-phenylindole) was produced from Roche Diagnostics (Germany). All PCR primers were purchased from Eurofins Genomics Inc. (Germany).
Cell cultures
Human alveolar epithelial progenitor cells were provided by Tohoku University and maintained in high- glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 20% fetal bovine serum (FBS). Human alveolar epithelial progenitor cells were incubated for indicated time or concentration of TTNPB or GW0742 in DMEM for and immunofluorescence staining cell count in 8well chamber (2 ×103/mL) and for western blot analysis in 6 well plate (5 × 103/mL), 0-48 hours in DMEM containing 1 μM GW0742 or ATRA for RT-PCR in 24 well plate (1 × 10104/mL).
Immunofluorescence staining and microscopy
Human alveolar epithelial progenitor cells were seeded at a concentration of 2 × 103/mL in DMEM containing 20% FBS and incubated for 6 days in the presence medium alone or medium containing TTNPB or GW0742 and medium was replaced new ligand-contained medium every 48 hour. The cells were fixed in 4% paraformaldehyde in PBS and blocked with 10% BSA in PBS and incubated overnight at 4°C with anti-SP-A antibody (1:200). After washing with PBS, the well was incubated 1.5 hour at room temperature with DAPI (20 μg/mL) and fluorescence conjugated antibodies, fluorescein isothiocyanate (FITC)-conjugated antibody (1:200) and washed with PBS-T. Determination of surface markers was performed by a direct immunofluorescence assay using the following samples were observed by fluorescent microscopy (BZ- 9000, KEYENCE, Japan).
Rate of positive cells was calculated by the number of FITC positive cells divided by DAPI positive cells and rate of positive cells in control group was regarded as normal speed of differentiation (100%).
Reverse transcription and RT-PCR
Total genes were extracted and clean up using RNAiso Plus (Takara, Japan). The isolated RNA was eluted in DEPC treated- Water (Nacalai, Japan) and assessed for quantity via UV spectrophotometry using NanoVue plus (GE Healthcare, UK) at 260/280 nm. Using PrimeScript® RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara, Japan) according to the manufacturer’s instructions, 20 ng total RNA was reverse transcribed to cDNA. The mRNA levels of the target and reference gene were measured using SYBR®Premix Ex Taq™ II (Tli RNaseH Plus) (Takara, Japan) and CFX Connect Real-Time PCR Detection System (Bio-Rad, USA). PCR cycling conditions included a 95°C heating step for 30 sec at the beginning of every run. The samples were then cycled at 95°C for 5 sec, annealed at 60°C for 30 sec. Optical data was collected after the annealing step. A melting curve was generated at the end of every run to ensure product uniformity. The primer sequences of ADRP [16], PPARβ/δ [17] and GAPDH [3] are shown in (Table 1). GAPDH was used as reference gene.
| Gene | Sequence | |
|---|---|---|
| GAPDH | Forward | 5’-GCACCGTCAAGGCTGAGAAC-3’ |
| GAPDH | Reverse | 5’-TGGTGAAGACGCCAGTGGA-3’ |
| ADRP | Forward | 5'-TGAGATGGCAGAGAACGGTGTG-3' |
| ADRP | Reverse | 5'-GGCATTGGCAACAATCTGAGT-3' |
| PPARβ/δ | Forward | 5'-TCTACAATGCCTACCTGAAAAACTTC-3' |
| PPARβ/δ | Reverse | 5'-ACAATGTCTCGATGTCGTGGATC-3' |
Table 1: Sequences for primers.
Electrophoresis and western blotting
Samples were collected with SDS buffer and boiled 3 min at 95°C. Reaction mixtures were separated on a 15% SDS gel with a 4.5% stacking gel at 200 volts. Proteins were transferred to polyvinylidene difluoride with a semi-dry electroblotter system (BioRad, USA) for 30 min at 10 volts. Subsequently, the membrane was blocked with 0.3% non-fat dry milk in Tris buffered saline TBS-0.1% Tween-20 (TBST) and incubated overnight at 4°C with the indicated primary antibody as follows: anti-SP-A antibody (1:1,000), or anti-actin antibody (1:1,000). After washing with TBST, the membrane was incubated 1 hour at room temperature with donkey anti-goat IgG-HRP antibody (1:25,000). Detection was performed using Immobilon ™ Western Chemiluminescent HRP Substrate (Millipore, USA) and following that user guide. Membranes were scanned and signals were analyzed with ChemiDoc™ Imaging Systems (BioRad, USA).
Small interfering RNA
Human alveolar epithelial progenitor cells were transfected with siRNA against PPARβ/δ and negative control (Ambion, Life Technologies,USA). Forty-eight hours prior to ligands treatment, cells in culture were transfected with 10 nM siRNA using Lipofectamine RNAiMAX reagent (Invitrogen, USA) and after starting exposure of ligand medium was replaced new ligand-contained medium every 48 hour. In order to examine the inhibitory effects of PPARβ/δ siRNA, mRNA was quantified by RT-PCR.
Statistics
Normally distributed data are presented as means ± standard deviation (SD). Student’s t-test was used when only two groups were compared. A p value < 0.01 (** or ??) or < 0.05 (* or ?) was considered statistically significant.
Results and Discussion
We have previously shown that ATRA induces the differentiation of human AEPCs to AT-II cells [3]; however, the mechanism by which ATRA induces AEPC differentiation remains unknown. In this study, we assessed the involvement of nuclear receptors RAR and PPARβ/δ in the differentiation of human AEPCs using the RAR-selective ligand TTNPB and the PPARβ/δ-selective ligand GW0742.
Effects of TTNPB and GW0742 on the differentiation of human AEPCs
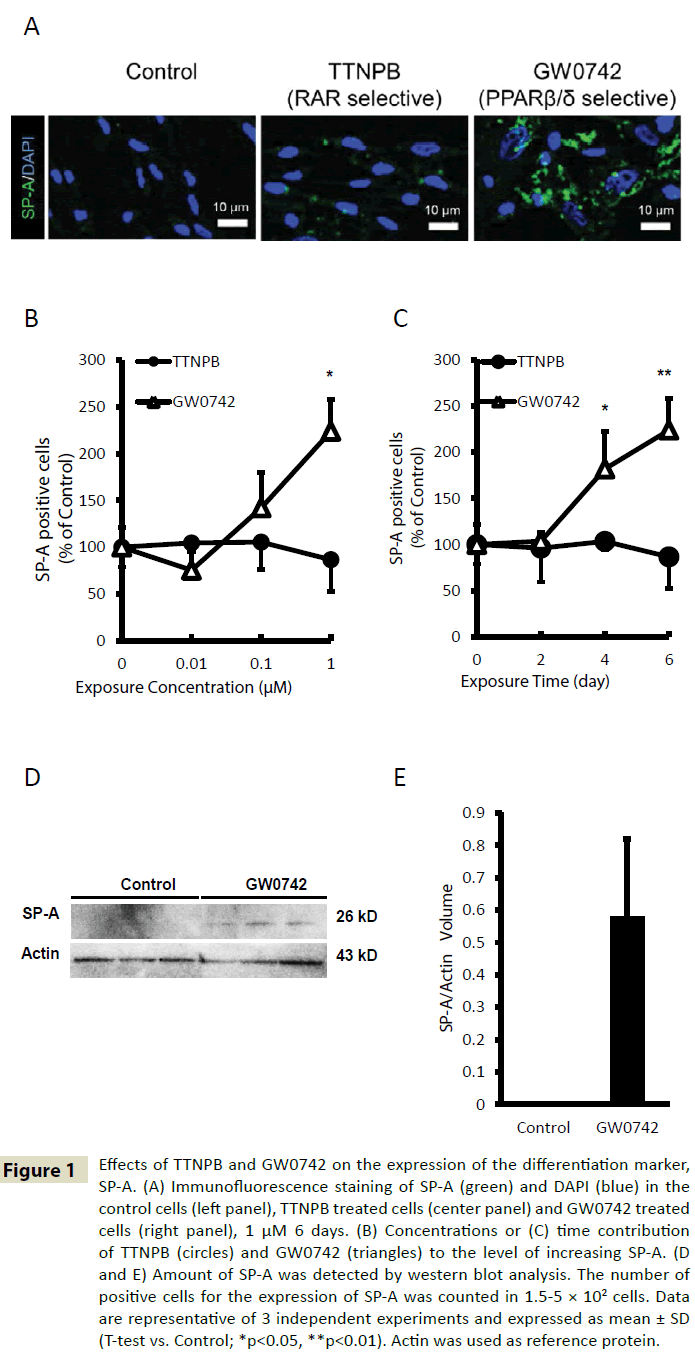
The level of differentiation of human AEPCs was evaluated using immunofluorescence to determine the levels of surfactant protein A (SP-A), a marker of AT-II cells. TTNPB did not induce differentiation of human AEPCs, but GW0742 could induce differentiation of human AEPCs into AT-II (Figure 1A). The SP-A levels after treatment with the indicated concentrations of TTNPB (0–1 μM) for the indicated time (0–6 days) was insignificant. However, treatment with GW0742 resulted in a significant dose-dependent (Figure 1B) and time-dependent (Figure 1C) increase in SP-A expression. The fraction of SP-A-positive cells present in human AEPCs treated with 1 μM GW0742 for 6 days was approximately 2.2-fold higher than the fraction present in the control group. According to the results of the western blot analysis, the SP-A levels in GW0742-treated cells also increased compared with that in the control cells (Figure 1D and 1E). The SP-A levels in extracts from control cells were below the limit of detection; however, a band corresponding to SP-A was detected in extracts from GW0742-treated cells. Thus, exposure of human AEPCs to 1 μM GW0742 for 6 days increases the rate of human AEPC differentiation 2.2-fold compared with the rate of differentiation of solvent-treated (0.1% DMSO, 6 days) control cells.
Figure 1: Effects of TTNPB and GW0742 on the expression of the differentiation marker, SP-A. (A) Immunofluorescence staining of SP-A (green) and DAPI (blue) in the control cells (left panel), TTNPB treated cells (center panel) and GW0742 treated cells (right panel), 1 μM 6 days. (B) Concentrations or (C) time contribution of TTNPB (circles) and GW0742 (triangles) to the level of increasing SP-A. (D and E) Amount of SP-A was detected by western blot analysis. The number of positive cells for the expression of SP-A was counted in 1.5-5 × 102 cells. Data are representative of 3 independent experiments and expressed as mean ± SD (T-test vs. Control; *p<0.05, **p<0.01). Actin was used as reference protein.
Effects of GW0742 and ATRA on mRNA expression of PPARβ/δ target genes
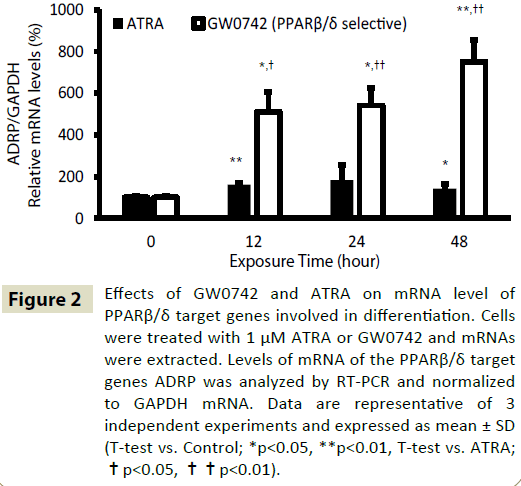
Treatment of human AEPCs with GW0742 and ATRA increased the expression of PPARβ/δ target gene mRNA (Figure 2). To quantify this effect, we selected adipose differentiation-related protein (ADRP), also known as PLIN2, as a representative PPARβ/δ target gene. The level of ADRP mRNA after treatment of human AEPCs with 1 μM ATRA or GW0742 for the indicated time (0–48 h) was evaluated by RT-PCR. Both ATRA and GW0742 significantly increased the level of ADRP mRNA in time-dependent manner compared with the control. Moreover, GW0742 treatment increased ADRP mRNA levels to a significantly greater extent than ATRA treatment. Treatment of human AEPCs with 1 μM GW0742 for 48 h increased ADRP mRNA levels 5.2-times than ATRA treatment under the same conditions (Figure 2). However, ATRA and GW0742 treatment did not influence the mRNA levels of PPARβ/δ or fatty acid binding protein 5 (FABP5) (data not shown). Reportedly, the levels of FABP5, a carrier of ATRA to PPARβ/δ nuclear receptor [4,18] are significantly decreased in the airway epithelial cells of smokers with COPD compared with those in smokers without COPD [19].
Figure 2: Effects of GW0742 and ATRA on mRNA level of PPARβ/δ target genes involved in differentiation. Cells were treated with 1 μM ATRA or GW0742 and mRNAs were extracted. Levels of mRNA of the PPARβ/δ target genes ADRP was analyzed by RT-PCR and normalized to GAPDH mRNA. Data are representative of 3 independent experiments and expressed as mean ± SD (T-test vs. Control; *p<0.05, **p<0.01, T-test vs. ATRA; ÃÆâÃâà âÃâÃÂp<0.05, ÃÆâÃâà âÃâÃÂÃÆâÃâà âÃâÃÂp<0.01).
In a study examining the ability of ATRA to activate PPARβ/δ in the human keratinocyte cell line HaCaT, ATRA signi?cantly upregulated ADRP [4]. The ADRP levels also increase during keratinocyte differentiation [10]. Fujino et al. demonstrated that human AEPCs in the extracellular matrix can be differentiated upon stimulation with keratinocyte growth factor, cAMP, or 3-isobutyl-1-methylxanthine. Using a microarray analysis, we recently showed that ATRA increased the expression of integrin and induced differentiation via phosphorylation signaling of AKT [3]. PPARβ/δ modulates Akt1 activation via transcriptional upregulation of integrin-linked kinase and PDK1. PDK1 causes higher Akt1 activity and results in increased keratinocyte differentiation in wound healing [20].
Effects of PPARβ/δ knockdown on the differentiation of human AEPCs
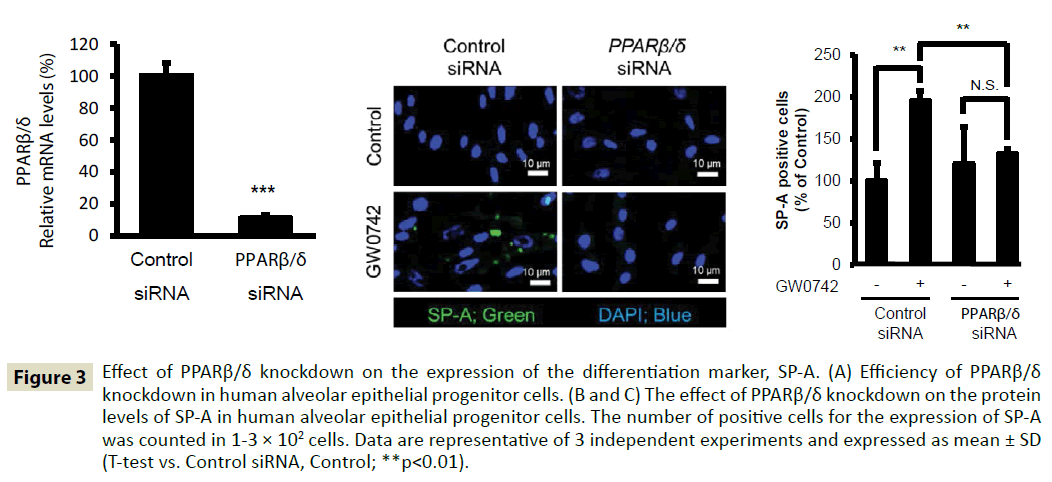
To evaluate the role played by PPARβ/δ nuclear receptor in the differentiation of human AEPCs, we treated human AEPCs with PPARβ/δ siRNA. Treatment with PPARβ/δ siRNA inhibited PPARβ/δ expression by 89% (Figure 3A). Treatment with PPARβ/δ siRNA significantly suppressed the differentiation of human AEPCs to AT-II cells induced by PPARβ/δ ligand GW0742 (Figure 3B and 3C). Therefore, PPARβ/δ knockdown suppresses the differentiation of human AEPCs to AT-II cells induced by GW0742 treatment.
Figure 3: Effect of PPARβ/δ knockdown on the expression of the differentiation marker, SP-A. (A) Efficiency of PPARβ/δ knockdown in human alveolar epithelial progenitor cells. (B and C) The effect of PPARβ/δ knockdown on the protein levels of SP-A in human alveolar epithelial progenitor cells. The number of positive cells for the expression of SP-A was counted in 1-3 × 102 cells. Data are representative of 3 independent experiments and expressed as mean ± SD (T-test vs. Control siRNA, Control; **p<0.01).
Human AEPCs, which originally express CD90, become cells that express SP-A after differentiation [2,3]. Because AT-II cells secrete SP-A, we used SP-A as a marker for AT-II cells. AT-II cells have a larger nucleus than human AEPCs and secrete a surfactant protein that reduces alveolar surface tension and maintains the shape of alveoli. Moreover, AT-II can differentiate to AT-I cells. AT-I cells cover majority of the surface area of the alveoli [21,22]. Therefore, it expected that the differentiation of human AEPCs to AT-II cells plays an important role in this aspect of alveolar repair.
Including other subtypes of PPAR, PPARα, PPARγ, and PPARβ/δ have been reported to be useful as therapeutic targets for lung inflammation because their ligands have potent anti-inflammatory activity [23-25]. However, PPARs have not been shown to have a role in regulating lung development [26]. In addition, there has been no description of the role for PPARβ/δ in modulating human lung cell development, although PPARβ/δ may have a role in the squamous differentiation process of tracheobronchial epithelial cells [27]. Therefore, this study is the ?rst to bring new insight to the differentiation of human AEPCs to alveolar epithelial cells induced by stimulation of PPARβ/δ. In addition, PPARβ/δ selective agonist, GW0742, could avoid cytotoxic [4] derived from stimulation to RAR, such as teratogenicity. Although further studies are required to establish the role of PPARβ/δ in alveolar Cells, this study indicates the possibility that PPARβ/δ agonists are useful as new therapeutic agents for pulmonary emphysema.
Acknowledgement
We thank Dr. Hiroshi Kubo (Tohoku University, Sendai, Japan) for provided human alveolar epithelial progenitor cells. This study was partially supported by the Grant-in-Aid for Research Activity Start-up [Michiko Horiguchi, 24890257], and the Grant-in Aid for Young Scientists (B) [Michiko Horiguchi, 25860029] from the Japan Society for the Promotion of Science.
Conflict of Interest
There are no conflicts of interest to declare.
References
- Barnes PJ(2000) Mechanisms in COPD: differences from asthma. Chest117:10s-14s.
- Fujino N, Kubo H, Suzuki T, Ota C, Hegab AE, et al. (2011) Isolation of alveolar epithelial type II progenitor cells from adult human lungs. Laboratory investigation: a journal of technical methods and pathology 91:363-378.
- Horiguchi M, Kojima H, Sakai H, Kubo H, Yamashita C (2014)Pulmonary administration of integrin-nanoparticles regenerates collapsed alveoli. Journal of controlled release: official journal of the Controlled Release Society 187:167-174.
- Schug TT, Berry DC, Shaw NS, Travis SN, Noy N (2007) Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell 129:723-733.
- Donato LJ, Suh JH, Noy N(2007) Suppression of mammary carcinoma cell growth by retinoic acid: the cell cycle control gene Btg2 is a direct target for retinoic acid receptor signaling. Cancer Res 67:609-615.
- Botling J, Castro DS, Oberg F, Nilsson K, Perlmann T (1997) Retinoic acid receptor/retinoid X receptor heterodimers can be activated through both subunits providing a basis for synergistic transactivation and cellular differentiation. The Journal of biological chemistry 272:9443-9449.
- Laurenzana A, Cellai C, Vannucchi AM, Pancrazzi A, Romanelli MN, et al. (2005) WEB-2086 and WEB-2170 trigger apoptosis in both ATRA-sensitive and -resistant promyelocytic leukemia cells and greatly enhances ATRA differentiation potential. Leukemia 19:390-395.
- Yao PL, Morales JL, Zhu B, Kang BH, Gonzalez FJ, et al. (2014) Activation of peroxisome proliferator-activated receptor-beta/delta (PPAR-beta/delta) inhibits human breast cancer cell line tumorigenicity. Molecular cancer therapeutics 13:1008-1017.
- Schmuth M, Haqq CM, Cairns WJ, Holder JC, Dorsam S, et al. (2004) Peroxisome proliferator-activated receptor (PPAR)-beta/delta stimulates differentiation and lipid accumulation in keratinocytes. The Journal of investigative dermatology 122:971-983.
- Chon SH, TannahillR, Yao X, Southall MD, Pappas A(2015) Keratinocyte differentiation and upregulation of ceramide synthesis induced by an oat lipid extract via the activation of PPAR pathways. Experimental dermatology 24:290-295.
- Dahlhoff M, Camera E, Picardo M, Zouboulis CC, Chan L, et al. (2013) PLIN2, the major perilipin regulated during sebocyte differentiation, controls sebaceous lipid accumulation in vitro and sebaceous gland size in vivo. BiochimBiophysActa1830: 4642-4649.
- Pignatello MA, Kauffman FC, Levin AA (1997) Multiple factors contribute to the toxicity of the aromatic retinoid, TTNPB (Ro 13-7410): binding affinities and disposition. Toxicology and applied pharmacology 142: 319-327.
- Klaholz BP, Mitschler A, Moras D (2000) Structural basis for isotype selectivity of the human retinoic acid nuclear receptor. Journal of molecular biology 302:155-170
- Sznaidman ML, Haffner CD, Maloney PR, Fivush A, Chao E, et al.(2003)Novel selective small molecule agonists for peroxisome proliferator-activated receptor delta (PPARdelta)--synthesis and biological activity. Bioorg Med ChemLett 13: 1517-1521.
- de Groot JC, Weidner C, Krausze J, Kawamoto K, Schroeder FC, et al. (2013) Structural characterization of amorfrutins bound to the peroxisome proliferator-activated receptor gamma. Journal of medicinal chemistry 56:1535-1543.
- Brown JD, Oligino E, Rader DJ, Saghatelian A, Plutzky J (2011) VLDL hydrolysis by hepatic lipase regulates PPARdelta transcriptional responses. PLoS One 6:e21209.
- Luo G, Shi Y, Zhang J, Mu Q, Qin L, et al. (2014) Palmitic acid suppresses apolipoprotein M gene expression via the pathway of PPARbeta/delta in HepG2 cells. BiochemBiophys Res Commun445:203-207.
- Morgan E, Kannan-Thulasiraman P, Noy N (2010) Involvement of Fatty Acid Binding Protein 5 and PPARbeta/delta in Prostate Cancer Cell Growth. PPAR research.
- Gally F, Chu HW, Bowler RP (2013) Cigarette smoke decreases airway epithelial FABP5 expression and promotes Pseudomonas aeruginosa infection. PLoS One 8: e51784.
- Di-Poi N, Tan NS, Michalik L, Wahli W, Desvergne B (2002) Antiapoptotic role of PPARbeta in keratinocytes via transcriptional control of the Akt1 signaling pathway. Molecular cell 10:721-733.
- Kubo H (2011) Molecular basis of lung tissue regeneration. General thoracic and cardiovascular surgery 59:231-244.
- Gotoh S, Ito I, Nagasaki T, Yamamoto Y, Konishi S, et al. (2014) Generation of alveolar epithelial spheroids via isolated progenitor cells from human pluripotent stem cells. Stem cell reports 3: 394-403.
- Haskova Z, Hoang B, Luo G, Morgan LA, Billin AN, et al. (2008) Modulation of LPS-induced pulmonary neutrophil infiltration and cytokine production by the selective PPARbeta/delta ligand GW0742. Inflammation research: official journal of the European Histamine Research Society57:314-321.
- Di Paola R, Crisafulli C, Mazzon E, Esposito E,Paterniti I, et al. (2010) GW0742, a high-affinity PPAR -beta/delta agonist, inhibits acute lung injury in mice. Shock 33:426-435.
- Galuppo M, Di Paola R, Mazzon E, Esposito E, Paterniti I, et al. (2010) GW0742, a high affinity PPAR-beta/delta agonist reduces lung inflammation induced by bleomycin instillation in mice. International journal of immunopathology and pharmacology 23: 1033-1046.
- Paola RD, Cuzzocrea S. (2007) Peroxisome proliferator-activated receptors and acute lung injury. PPAR research 2007: 63745.
- Matsuura H, Adachi H, Smart RC, Xu X, Arata J, et al. (1999) Correlation between expression of peroxisome proliferator-activated receptor beta and squamous differentiation in epidermal and tracheobronchial epithelial cells. Molecular and cellular endocrinology 147: 85-92.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences